Machine Learning and Atom-Based Quadratic Indices for Proteasome Inhibition Prediction
http://repository.vnu.edu.vn/handle/VNU_123/11510
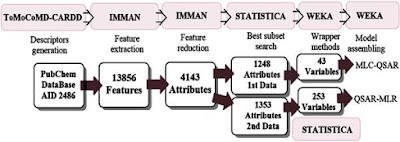
The atom-based quadratic indices are used in this work together with some machine learning techniques that includes: support vector machine, artificial neural network, random forest and k-nearest neighbor.
This methodology is used for the development of two quantitative structure-activity relationship (QSAR) studies for the prediction of proteasome inhibition.
A first set consisting of active and non-active classes was predicted with model performances above 85% and 80% in training and validation series, respectively. These results provided new approaches on proteasome inhibitor identification encouraged by virtual screenings procedures
The atom-based quadratic indices are used in this work together with some machine learning techniques that includes: support vector machine, artificial neural network, random forest and k-nearest neighbor.
This methodology is used for the development of two quantitative structure-activity relationship (QSAR) studies for the prediction of proteasome inhibition.
A first set consisting of active and non-active classes was predicted with model performances above 85% and 80% in training and validation series, respectively. These results provided new approaches on proteasome inhibitor identification encouraged by virtual screenings procedures
| Title: | Machine Learning and Atom-Based Quadratic Indices for Proteasome Inhibition Prediction |
| Authors: | Le, Thi Thu Huong |
| Keywords: | Atom-based quadratic index;Classification and regression model;Machine learning;Proteasome inhibition;QSAR;TOMOCOMD-CARDD software |
| Issue Date: | 2015 |
| Publisher: | Mol2Net |
| Abstract: | The atom-based quadratic indices are used in this work together with some machine learning techniques that includes: support vector machine, artificial neural network, random forest and k-nearest neighbor. This methodology is used for the development of two quantitative structure-activity relationship (QSAR) studies for the prediction of proteasome inhibition. A first set consisting of active and non-active classes was predicted with model performances above 85% and 80% in training and validation series, respectively. These results provided new approaches on proteasome inhibitor identification encouraged by virtual screenings procedures |
| URI: | http://repository.vnu.edu.vn/handle/VNU_123/11510 |
| ISSN: | 1422-0067 |
| Appears in Collections: | SMP - Papers / Tham luận HN-HT |


Nhận xét
Đăng nhận xét